Citations & Publications
Of 43 papers, those that were co-authored are marked as such:

Martin LE, Barr JS, Cartailler JP, Shrestha S, Estévez-Lao TY, Hillyer JF. Warmer temperature accelerates senescence by modifying the aging-dependent changes in the mosquito transcriptome, altering immunity, metabolism, and DNA repair. Immun Ageing. 2025 Dec 13;23(1):1. doi: 10.1186/s12979-025-00551-7. PMID: 41390729; PMCID: PMC12781269. Read it
(CDS contribution: RNA-Seq Analysis)
(CDS contribution: RNA-Seq Analysis)
Luo X, Huang Y, Zeng H, Tao Y, Bao X, Feng F, Hopkirk AL, Pham T, Bate TSR, Saunders DC, Orchard P, Robertson C, Shrestha S, Spraggins JM, Cartailler JP, Parker SCJ, Brissova M, Liu J. CelLink: integrating single-cell multi-omics data with weak feature linkage and imbalanced cell populations. Nucleic Acids Res. 2025 Nov 26;53(22):gkaf1270. doi: 10.1093/nar/gkaf1270. PMID: 41335468; PMCID: PMC12818349. Read it
(CDS contribution: software-engineering-programming)
(CDS contribution: software-engineering-programming)
Dynamic Ca2+-Dependent Transcription Links Metabolic Stress to Impaired β-Cell Identity. Anna B. Osipovich, Matthew T. Dickerson, Jean-Philippe Cartailler, Shristi Shrestha, Nicole M. Wright, David A. Jacobson, Mark A. Magnuson; Diabetes 2025; db241141. https://doi.org/10.2337/db24-1141 Read it
(CDS contribution: RNA-Seq, WGCNA, temporal analysis)
(CDS contribution: RNA-Seq, WGCNA, temporal analysis)

E3 ligase substrate adaptor SPOP fine-tunes the UPR of pancreatic β cells. Oguh AU, Haemmerle MW, Sen S, Rozo AV, Shrestha S, Cartailler JP, Fazelinia H, Ding H, Preza S, Yang J, Yang X, Sussel L, Alvarez-Dominguez JR, Doliba N, Spruce LA, Arrojo E Drigo R, Stoffers DA. Genes Dev. 2024 Dec 30. doi: 10.1101/gad.352010.124. Epub ahead of print. PMID: 39797759. Read it
(CDS contribution: pySCENIC, regulons, single cell transcriptomics)
(CDS contribution: pySCENIC, regulons, single cell transcriptomics)
Caloric restriction promotes beta cell longevity and delays aging and senescence by enhancing cell identity and homeostasis mechanisms. Dos Santos C, Shrestha S, Cottam M, Perkins G, Lev-Ram V, Roy B, Acree C, Kim KY, Deerinck T, Cutler M, Dean D, Cartailler JP, MacDonald PE, Hetzer M, Ellisman M, Drigo RAE. Nat Commun 15, 9063 (2024). https://doi.org/10.1038/s41467-024-53127-2 Read it
(CDS contribution: Multi-ome data processing and analysis, pySCENIC regulon enrichment analysis)
(CDS contribution: Multi-ome data processing and analysis, pySCENIC regulon enrichment analysis)

Disruption of nucleotide biosynthesis reprograms mitochondrial metabolism to inhibit adipogenesis. Pinette JA, Myers JW, Park WY, Bryant HG, Eddie AM, Wilson GA, Montufar C, Shaikh Z, Vue Z, Nunn ER, Bessho R, Cottam MA, Haase VH, Hinton AO, Spinelli JB, Cartailler JP, Zaganjor E. Journal of Lipid Research Read it
(CDS contribution: RNA-Seq, functional enrichment analysis, data deposition)
(CDS contribution: RNA-Seq, functional enrichment analysis, data deposition)

Knockdown of microglial iron import gene, Slc11a2, worsens cognitive function and alters microglial transcriptional landscape in a sex-specific manner in the APP/PS1 model of Alzheimer's disease. Robertson KV, Rodriguez AS, Cartailler JP, Shrestha S, Schleh MW, Schroeder KR, Valenti AM, Kramer AT, Harrison FE, Hasty AH. J Neuroinflammation. 2024 Sep 27;21(1):238. doi: 10.1186/s12974-024-03238-w. PMID: 39334471; PMCID: PMC11438269. Read it
(CDS contribution: RNA-seq experiments)
(CDS contribution: RNA-seq experiments)

CNTools: A computational toolbox for cellular neighborhood analysis from multiplexed images. Tao Y, Feng F, Luo X, Reihsmann CV, Hopkirk AL, Cartailler JP, Brissova M, Parker SCJ, Saunders DC, Liu J. PLoS Comput Biol. 2024 Aug 28;20(8):e1012344. doi: 10.1371/journal.pcbi.1012344. PMID: 39196899. Read it
(CDS contribution: Testing and validation of the CNTools package)
(CDS contribution: Testing and validation of the CNTools package)

Transcriptomic Plasticity of the Circadian Clock in Response to Photoperiod: A Study in Male Melatonin-Competent Mice. Cox OH, Gianonni-Guzmán MA, Cartailler J-P, Cottam MA, McMahon DG. Journal of Biological Rhythms. 2024;0(0). Read it
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression, differential circadian analysis, Shiny app development)
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression, differential circadian analysis, Shiny app development)

Rescue of impaired blood-brain barrier in tuberous sclerosis complex patient derived neurovascular unit. Brown JA, Faley SL, Judge M, Ward P, Ihrie RA, Carson R, Armstrong L, Sahin M, Wikswo JP, Ess KC, Neely MD. J Neurodev Disord. 2024 May 23;16(1):27. doi: 10.1186/s11689-024-09543-y. PMID: 38783199; PMCID: PMC11112784. Read it
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)

The MODY-associated KCNK16 L114P mutation increases islet glucagon secretion and limits insulin secretion resulting in transient neonatal diabetes and glucose dyshomeostasis in adults. Nakhe AY, Dadi PK, Kim J, Dickerson MT, Behera S, Dobson JR, Shrestha S, Cartailler JP, Sampson L, Magnuson MA, Jacobson DA. Elife. 2024 May 3;12:RP89967. doi: 10.7554/eLife.89967. PMID: 38700926; PMCID: PMC11068355. Read it
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)

Differential Signaling Effects of Escherichia Coli and Staphylococcus Aureus in Human Whole Blood Indicate Distinct Regulation of the NRF2 Pathway.. Pourquoi A, Miller MR, Koch SR, Boyle K, Surratt V, Nguyen H, Panja S, Cartailler JP, Shrestha S, Stark RJ. Shock 61(4):p 557-563, April 2024. | DOI: 10.1097/SHK.0000000000002305 Read it
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)

From goal to outcome: Analyzing the progression of biomedical sciences PhD careers in a longitudinal study using an expanded taxonomy. Brown AM, Meyers LC, Varadarajan J, Ward NJ, Cartailler JP, Chalkley RG, Gould KL, Petrie KA. FASEB BioAdvances. 2023; 00: 1-26. doi:10.1096/fba.2023-00072 Read it
(CDS contribution: Database architecture and development, ETL, publication/software engineering, documentation, infrastructure)
(CDS contribution: Database architecture and development, ETL, publication/software engineering, documentation, infrastructure)

Deletion of Ascl1 in pancreatic β-cells improves insulin secretion, promotes parasympathetic innervation, and attenuates dedifferentiation during metabolic stress. Osipovich AB, Zhou FY, Chong JJ, Trinh LT, Cottam MA, Shrestha S, Cartailler JP, Magnuson MA. Mol Metab. 2023 Sep 26:101811. doi: 10.1016/j.molmet.2023.101811. Epub ahead of print. PMID: 37769990. Read it
(CDS contribution: Bioinformatics, RNA-Seq)
(CDS contribution: Bioinformatics, RNA-Seq)

A framework to identify functional interactors that contribute to disrupted early retinal development in Vsx2 ocular retardation J mice. Leung AM, Rao MB, Raju N, Chung M, Klinger A, Rowe DJ, Li X, Levine EM. Dev Dyn. 2023 Nov;252(11):1338-1362. doi: 10.1002/dvdy.629. Epub 2023 Jun 1. PMID: 37259952; PMCID: PMC10689574. Read it
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)

The human α cell in health and disease. Pettway YD, Saunders DC, Brissova M. J Endocrinol. 2023 Jun 19;258(1):e220298. doi: 10.1530/JOE-22-0298. PMID: 37114672; PMCID: PMC10428003. Read it
(CDS contribution: RNA-Seq, primate data)
(CDS contribution: RNA-Seq, primate data)

ZFP92, a KRAB domain zinc finger protein enriched in pancreatic islets, binds to B1/Alu SINE transposable elements and regulates retroelements and genes. Osipovich AB, Dudek KD, Trinh LT, Kim LH, Shrestha S, Cartailler JP, Magnuson MA. PLoS Genet. 2023 May 8;19(5):e1010729. doi: 10.1371/journal.pgen.1010729. PMID: 37155670. Read it
(CDS contribution: Bulk RNA-Seq analysis, transcriptomics, transposable elements, data visualization, enrichment analysis)
(CDS contribution: Bulk RNA-Seq analysis, transcriptomics, transposable elements, data visualization, enrichment analysis)

Single-cell RNA sequencing of Sox17-expressing lineages reveals distinct gene regulatory networks and dynamic developmental trajectories. Stem Cells. Trinh LT, Osipovich AB, Liu B, Shrestha S, Cartailler JP, Wright CVE, Magnuson MA. 2023 Apr 21:sxad030. doi: 10.1093/stmcls/sxad030. Epub ahead of print. PMID: 37085274. Read it
(CDS contribution: Bioinformatics, pySCENIC, single cell transcriptomics, computational analysis)
(CDS contribution: Bioinformatics, pySCENIC, single cell transcriptomics, computational analysis)

Protein kinase D1 (Prkd1) deletion in brown adipose tissue leads to altered myogenic gene expression after cold exposure, while thermogenesis remains intact. Crowder, MK, Shrestha, S, Cartailler, JP, & Collins, S (2023). Physiological Reports, 11, e15576. https://doi.org/10.14814/phy2.15576 Read it
(CDS contribution: Bioinformatics, bulk RNA-Seq analysis, transcriptomics, data visualization, enrichment analysis)
(CDS contribution: Bioinformatics, bulk RNA-Seq analysis, transcriptomics, data visualization, enrichment analysis)

Human pancreatic capillaries and nerve fibers persist in type 1 diabetes despite beta cell loss. Richardson TM, Saunders DC, Haliyur R, Shrestha S, Cartailler JP, Reinert RB, Petronglo J, Bottino R, Aramandla R, Bradley AM, Jenkins R, Phillips S, Kang H; Human Pancreas Analysis Program; Caidedo A, Powers AC, Brissova M. Am J Physiol Endocrinol Metab. 2023 Jan 25. doi: 10.1152/ajpendo.00246.2022. Epub ahead of print. PMID: 36696598. Read it
(CDS contribution: Single cell transcriptomics, computational analysis)
(CDS contribution: Single cell transcriptomics, computational analysis)

Lhx2 is a progenitor-intrinsic modulator of Sonic Hedgehog signaling during early retinal neurogenesis. Li X, Gordon PJ, Gaynes JA, Fuller AW, Ringuette R, Santiago CP, Wallace V, Blackshaw S, Li P, Levine EM. Elife. 2022 Dec 2;11:e78342. doi: 10.7554/eLife.78342. PMID: 36459481; PMCID: PMC9718532. Read it
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)
(CDS contribution: Bioinformatics, RNA-Seq, differential gene expression)

Aging compromises human islet beta cell function and identity by decreasing transcription factor activity and inducing ER stress. Shrestha S, Erikson G, Lyon J, Spigelman AF, Bautista A, Manning Fox JE, Dos Santos C, Shokhirev M, Cartailler JP, Hetzer MW, MacDonald PE, Arrojo E Drigo R. Sci Adv. 2022 Oct 7;8(40):eabo3932. doi: 10.1126/sciadv.abo3932. Epub 2022 Oct 5. PMID: 36197983; PMCID: PMC9534504. Read it
(CDS contribution: Single cell transcriptomics, pySCENIC, computational analysis)
(CDS contribution: Single cell transcriptomics, pySCENIC, computational analysis)

Distinct Patterns of Clonal Evolution Drive Myelodysplastic Syndrome Progression to Secondary Acute Myeloid Leukemia. Guess T, Potts CR, Bhat P, Cartailler JA, Brooks A, Holt C, Yenamandra A, Wheeler FC, Savona MR, Cartailler JP, Ferrell PB. Blood Cancer Discov. 2022 Jul 6;3(4):316-329. doi: 10.1158/2643-3230.BCD-21-0128. Read it
(CDS contribution: Bioinformatics, single cell transcriptomics, data visualization)
(CDS contribution: Bioinformatics, single cell transcriptomics, data visualization)
Human iPSC-derived cerebral organoids model features of Leigh syndrome and reveal abnormal corticogenesis. Romero-Morales AI, Robertson GL, Rastogi A, Rasmussen ML, Temuri H, McElroy GS, Chakrabarty RP, Hsu L, Almonacid PM, Millis BA, Chandel NS, Cartailler JP, Gama V. Development. 2022 Oct 15;149(20):dev199914. doi: 10.1242/dev.199914. Epub 2022 Jul 6. Read it
(CDS contribution: Bioinformatics, data visualization)
(CDS contribution: Bioinformatics, data visualization)

Isocitrate dehydrogenase mutations are associated with altered IL-1β responses in acute myeloid leukemia. Sunthankar KI, Jenkins MT, Cote CH, Patel SB, Welner RS, Ferrell PB. Leukemia. 2022 Apr;36(4):923-934. doi: 10.1038/s41375-021-01487-9. Epub 2021 Dec 2. PMID: 34857894; PMCID: PMC9066619. Read it
(CDS contribution: Single cell transcriptomics)
(CDS contribution: Single cell transcriptomics)

Influence of basal media composition on barrier fidelity within human pluripotent stem cell-derived blood-brain barrier models. Neal EH, Katdare KA, Shi Y, Marinelli NA, Hagerla KA, Lippmann ES. J Neurochem. 2021 Dec;159(6):980-991. doi: 10.1111/jnc.15532. Epub 2021 Nov 18. PMID: 34716922; PMCID: PMC8688328. Read it
(CDS contribution: Discussion about RNA-Seq experimental design)
(CDS contribution: Discussion about RNA-Seq experimental design)

Regulation and Functional Consequences of mGlu4 RNA Editing Hofmann CS, Carrington S, Keller AN, Gregory KJ, Niswender CM. RNA. 2021 Oct;27(10):1220-1240. doi: 10.1261/rna.078729.121 Read it
(CDS contribution: Bioinformatics of RNA editing events from NGS FastQ files)
(CDS contribution: Bioinformatics of RNA editing events from NGS FastQ files)

Mouse models of patent ductus arteriosus (PDA) and their relevance for human PDA. Yarboro MT, Gopal SH, Su RL, Morgan TM, Reese J. Dev Dyn. 2021 Aug 4. doi: 10.1002/dvdy.408. Read it
(CDS contribution: Protein-protein interaction analysis, data interpretation, and figure construction)
(CDS contribution: Protein-protein interaction analysis, data interpretation, and figure construction)

Insm1, Neurod1, and Pax6 promote murine pancreatic endocrine cell development through overlapping yet distinct RNA transcription and splicing programs. Dudek KD, Osipovich AB, Cartailler JP, Gu G, Magnuson MA. G3 Genes|Genomes|Genetics, 2021 Read it
(CDS contribution: Bulk RNA-Seq analysis, RNA alternate splicing analysis)
(CDS contribution: Bulk RNA-Seq analysis, RNA alternate splicing analysis)

Combinatorial transcription factor profiles predict mature and functional human islet α and β cells. Shrestha S, Saunders DC, Walker JT, Camunas-Soler J, Dai XQ, Haliyur R, Aramandla R, Poffenberger G, Prasad N, Bottino R, Stein R, Cartailler JP, Parker SC, MacDonald PE, Levy SE, Powers AC, Brissova M. JCI Insight. 2021 Aug 24:151621. doi: 10.1172/jci.insight.151621. Read it
(CDS contribution: Single cell RNA-Seq analysis)
(CDS contribution: Single cell RNA-Seq analysis)

Regulation of RNA editing by intracellular acidification. Malik TN, Doherty EE, Gaded VM, Hill TM, Beal PA, Emeson RB. Nucleic Acids Research, 2021 Read it
(CDS contribution: Data processing and analysis of RNA editing sites)
(CDS contribution: Data processing and analysis of RNA editing sites)

A developmental lineage-based gene co-expression network for mouse pancreatic β-cells reveals a role for Zfp800 in pancreas development. Osipovich AB, Dudek KD, Greenfest-Allen E, Cartailler JP, Manduchi E, Potter Case L, Choi E, Chapman AG, Clayton HW, Gu G, Stoeckert CJ Jr, Magnuson MA. Development. 2021 Mar 2:dev.196964. doi: 10.1242/dev.196964. Read it
(CDS contribution: Network analysis, web app development)
(CDS contribution: Network analysis, web app development)

Pancreatlas™: applying an adaptable framework to map the human pancreas in health and disease. Saunders DC, Messmer J, Kusmartseva I, Beery ML, Yang M, Atkinson MA, Powers AC, Cartailler JP, Brissova M. Patterns (N Y). 2020 Oct 5;1(8):100120. doi: 10.1016/j.patter.2020.100120. Read it
(CDS contribution: Informatics, imaging, infrastructure, programming)
(CDS contribution: Informatics, imaging, infrastructure, programming)

Quantitative Analysis of Adenosine-to-Inosine RNA Editing. Malik TN, Cartailler JP, Emeson RB. Methods Mol Biol. 2021;2181:97-111. doi:10.1007/978-1-0716-0787-9_7 Read it
(CDS contribution: Bioinformatics)
(CDS contribution: Bioinformatics)
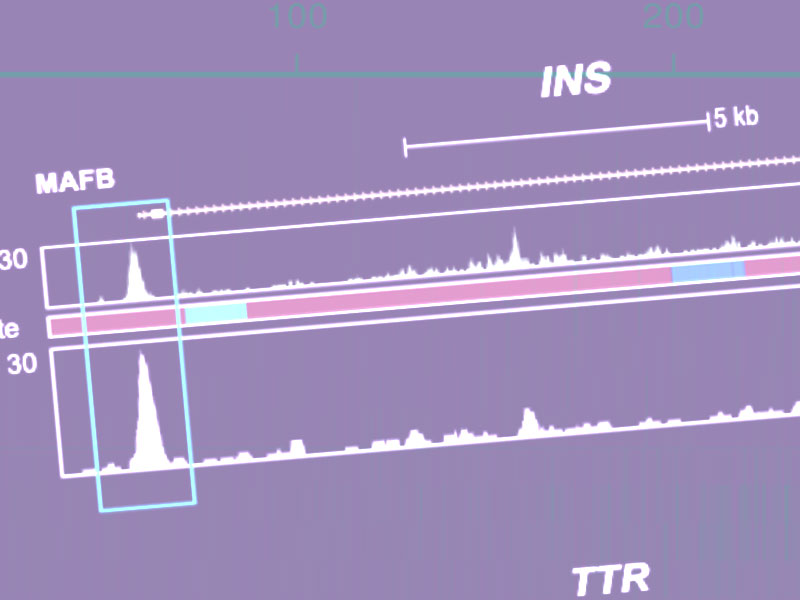
Loss of the transcription factor MAFB limits β-cell derivation from human PSCs Russell R, Carnese PP, Hennings TG, Walker EM, Russ HA, Liu JS, Giacometti S, Stein R & Hebrok M. Nature Communications. 2020 Read it
(CDS contribution: ChIP-Seq Analysis)
(CDS contribution: ChIP-Seq Analysis)

Excitotoxicity and overnutrition additively impair metabolic function and identity of pancreatic β-cells Osipovich AB, Stancill JS, Cartailler JP, Dudek KD, Magnuson MA. Diabetes. 2020 Jul;69(7):1476-1491. doi: 10.2337/db19-1145 Read it
(CDS contribution: Bioinformatics, RNA-Seq data processing and analysis, functional gene enrichment analysis)
(CDS contribution: Bioinformatics, RNA-Seq data processing and analysis, functional gene enrichment analysis)

Human iPSC-derived cerebral organoids model features of Leigh Syndrome and reveal abnormal corticogenesis Alejandra I. Romero-Morales, Anuj Rastogi, Hoor Temuri, Megan L. Rasmussen, Gregory Scott McElroy, Lawrence Hsu, Paula M. Almonacid, Bryan A. Millis, Navdeep S Chandel, Jean-Phillippe Cartailler, Vivian Gama. BioRxiv. 2020. doi: https://doi.org/10.1101/2020.04.21.054361 Read it
(CDS contribution: Bioinformatics, variant calling analysis, visualization, enrichment analysis)
(CDS contribution: Bioinformatics, variant calling analysis, visualization, enrichment analysis)

Transgene-associated human growth hormone expression in pancreatic β-cells impairs identification of sex-based gene expression differences. Stancill JS, Osipovich AB, Cartailler JP, Magnuson MA. Am J Physiol Endocrinol Metab. 2019 Feb 1;316(2):E196-E209. doi: 10.1152/ajpendo.00229.2018 Read it
(CDS contribution: Bulk RNA-Seq data processing and analysis)
(CDS contribution: Bulk RNA-Seq data processing and analysis)

iterativeWGCNA: iterative refinement to improve module detection from WGCNA co-expression networks. Greenfest-Allen E, Cartailler JP, Magnuson MA, Stoeckert CJ. BioRxiv. 2017. doi: https://doi.org/10.1101/234062 Read it
(CDS contribution: Method validation and application to new datasets)
(CDS contribution: Method validation and application to new datasets)

Chronic β-Cell Depolarization Impairs β-Cell Identity by Disrupting a Network of Ca2+-Regulated Genes. Stancill JS, Cartailler JP, Clayton HW, O'Connor JT, Dickerson MT, Dadi PK, Osipovich AB, Jacobson DA, Magnuson MA. Diabetes. 2017 Aug;66(8):2175-2187. doi: 10.2337/db16-1355 Read it
(CDS contribution: Bulk RNA-Seq data processing and analysis)
(CDS contribution: Bulk RNA-Seq data processing and analysis)

New ideas connecting the cell cycle and pancreatic endocrine-lineage specification. Bechard ME, Wright CV. Cell Cycle. 2017 Feb 16;16(4):301-303. doi: 10.1080/15384101.2016.1256149 Read it
(CDS contribution: Figure 1 illustration)
(CDS contribution: Figure 1 illustration)

Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Osipovich AB, Gangula R, Vianna PG, Magnuson MA. Development. 2016 Dec 15;143(24):4595-4607 Read it
(CDS contribution: Bulk RNA-Seq data processing and analysis)
(CDS contribution: Bulk RNA-Seq data processing and analysis)
Research resource: dkCOIN, the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) consortium interconnectivity network: a pilot program to aggregate research resources generated by multiple research consortia. McKenna NJ, Howard CL, Aufiero M, Easton-Marks J, Steffen DL, Becnel LB, Magnuson MA, McIndoe RA, Cartailler JP. Mol Endocrinol. 2012 Oct;26(10):1675-81 Read it
(CDS contribution: Informatics, strategy, leadership)
(CDS contribution: Informatics, strategy, leadership)
Bytes
Data Repositories
- E-MTAB-4726 (ArrayExpress)
- E-MTAB-5189 (ArrayExpress)
- E-MTAB-6329 (ArrayExpress)
- E-MTAB-6615 (ArrayExpress)
- E-MTAB-8643 (ArrayExpress)
- E-MTAB-8612 (ArrayExpress)
- E-MTAB-8802 (ArrayExpress)
- PRJNA626388 (NCBI)
- 7311202 (Zenodo)
Code & Data Repositories
Published code, apps and data:
- Circadian Photoperiod Seq: SCN Gene Rhythms and Expression Browser
- Code - A developmental lineage-based gene co-expression network for mouse pancreatic beta-cells
- Code - iterativeWGCNA: a WGCNA extension
- Code - Modeling the effect of Leigh syndrome-associated mutations using human cerebral organoids
- Code - Freezer configuration generator for Core Informatics LIMS (Python)
- Code - Generate Leaflet-based heatmap from collaborator-location data
- Code - Single cell Islets - R Shiny App
- Code - FFIND is a generalized framework for connecting to managed data and metadata from diverse sources and making it accessible via an integrated web application that promotes the exploration and discovery of the data.
- App - Single cell gene expression atlas of human pancreatic islets
- Code - Aging compromises human islet beta cell function and identity by decreasing transcription factor activity and inducing ER stress
- Data - Aging compromises human islet beta cell function and identity by decreasing transcription factor activity and inducing ER stress
